R Clinical
Our mission is to provide R Clinical Programming Services to Sponsors. Several key features of our platform enable us to deliver a superior product, such as using R packages with good life cycle management, a reproducible environment in R Studio, CDISC metadata and standards, automated scripts for the production of TLFs and Reports, and ensuring good QA/QC through SOPs, templates, and independent programming (i.e., reproducing the SDTM datasets with SAS).
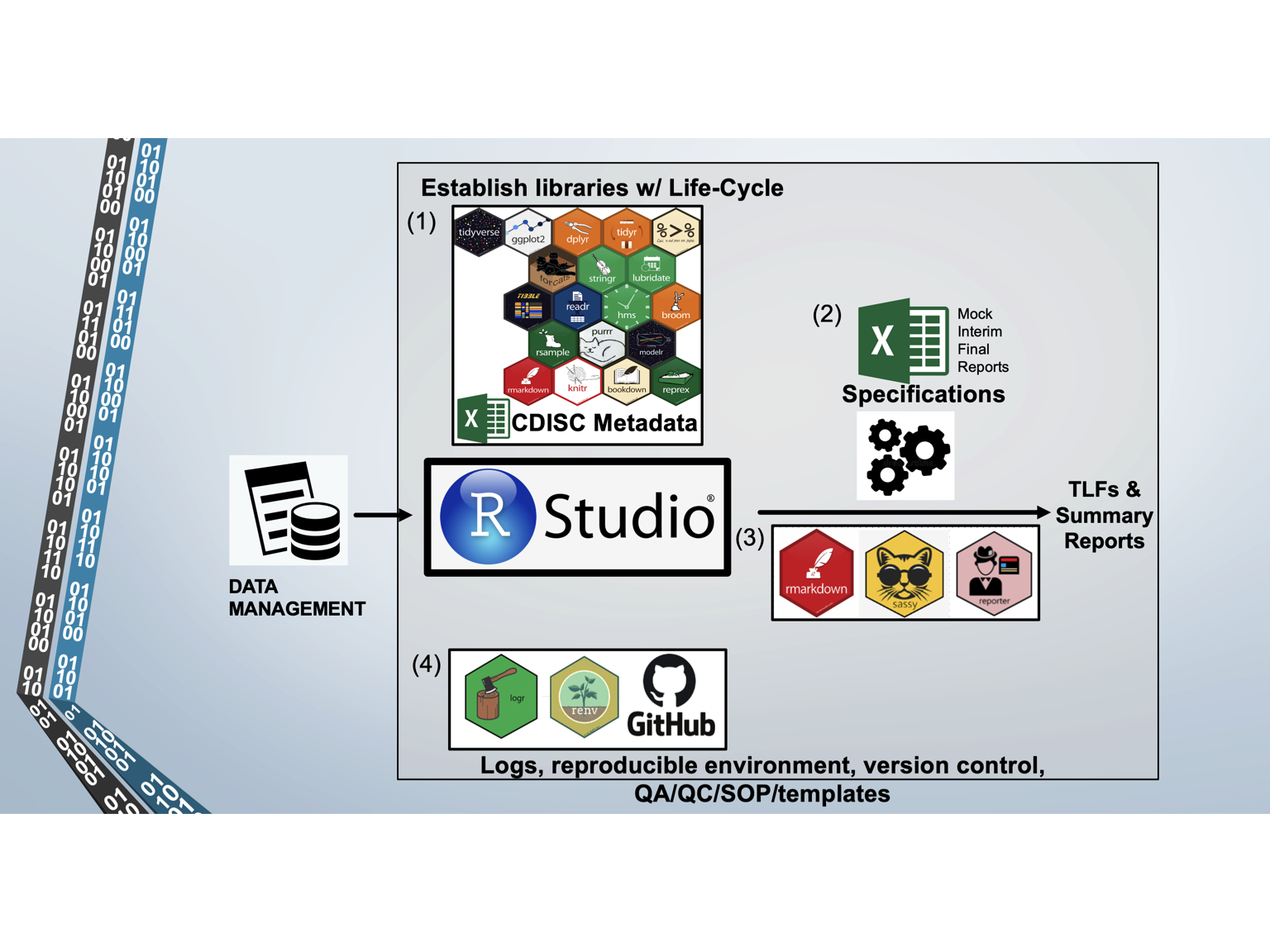
- R scripts
- path.R script establishes pathways and custom functions
- cleansdtm.R cleans raw trial data and maps to SDTM standards
- cleanadam.R maps SDTM datasets to ADAM standards/datasets
The path.R script contains thousands of lines of custom functions developed by Data InDeed to help stream line the production of SDTM and ADAM datasets.
For each trial, the raw data is cleaned and mapped to SDTM standards using the cleansdtm.R script. Based on CDISC metadata and standards, the script produces all SDTM datasets in CSV and XPT (SAS transport) extensions. The CSV file enables Sponsors to use the mapped data for additional analyses. The cleansdtm.R script is the most extensively coded script (several thousand lines) depending on the degree to which the raw data is already mapped to CDISC standards on the case report forms and in the clinical database. We have experience mapping raw data from RedCap, Castor, Medrio, and Oracle-based custom databases.
The cleanadam.R script generates all of the ADAM datasets for efficacy, safety, medical histories, concomitant medications, adverse events, and subject level data. The cleanadam.R script is several hundreds of lines of code; since it relies on mapping standard SDTM datasets, the scripts change little between studies.
All of the R code is fully documented and written in a SAS-friendly manner so that other SAS programmers can easily understand the R code.
- The specifications.xlsx file controls:
- What programs (R scripts) are sourced (i.e., an efficacy table)
- What and how variables are selected, filtered, arranged
- What parameters are used in the listing, table or figure
- What titles, subtitles, notes
- Where the outputs are saved
- Where the log files are saved
Each analysis is automated through the use of a specification.xlxs file, which defines all the variables for data presentation. When TLF and/or Reporting Scripts are run, all of the variables are reported in the scripts (which originated from the specification.xlxs file) so that complete documentation is logged. The specification.xlxs is a powerful way to make small adjustments to the TLFs (i.e., drop a subject from the analysis; change the population being analyzed, define the output name for deliverable).
- Rmarkdown, sassy, reporter
- Creates TLFs and Reports
- Formats
- Consistency
- Github, logr, renv provide
- Reproducibility
- Traceability
- Auditability
We also leverage Reproducible Research, which…
ties specific instructions to data analysis and experimental data so that scholarship can be recreated, better understood and verified. Packages in R for this purpose can be split into groups for: literate programming, package reproducibility, code/data formatting tools, format convertors, and object caching. The primary way that R facilitates reproducible research is using a document that is a combination of content and data analysis code. The Sweave function (in the base R utils package) and the knitr package can be used to blend the subject matter and R code so that a single document defines the content and the analysis.
As a result, we have developed custom workflows and can develop new custom workflows for your research needs. A great graphic that illustrates this can be found at the Aberdeen Study Group. We can develop a similar workflow to provide a solution to your data analysis problems!
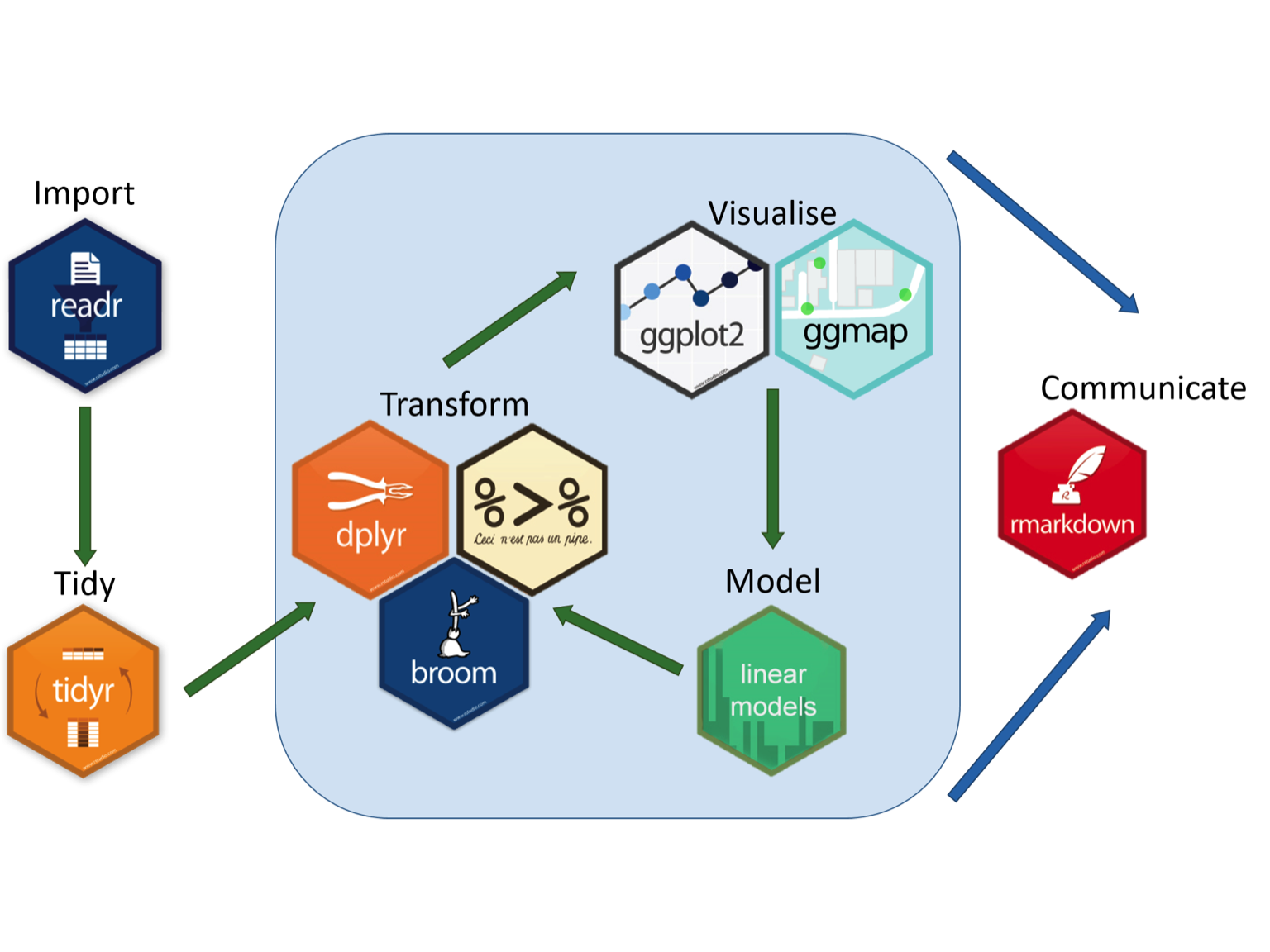
We can also use RMarkdown to support your publication needs including support with artwork preparation, pre-submission review, journal selection, journal submission, plagiarism check, rapid technical review, and re-submission support.
Other key support includes:
- Ensure that your text, figures, tables, and references are all formatted per the “instructions to author” guidelines.
- Science editing service to address language, structure, overall logic, flow and strength of your research.
